Data from: Multiscale Estimation of Binding Kinetics Using Brownian Dynamics, Molecular Dynamics and Milestoning
Data from: Multiscale Estimation of Binding Kinetics Using Brownian Dynamics, Molecular Dynamics and Milestoning
About this collection
- Extent
-
1 digital object.
- Cite This Work
-
Votapka, Lane W.; Amaro, Rommie E. (2015). Data from: Multiscale estimation of binding kinetics using Brownian dynamics, molecular dynamics and milestoning. UC San Diego Library Digital Collections. https://doi.org/10.6075/J02Z13F5
- Description
-
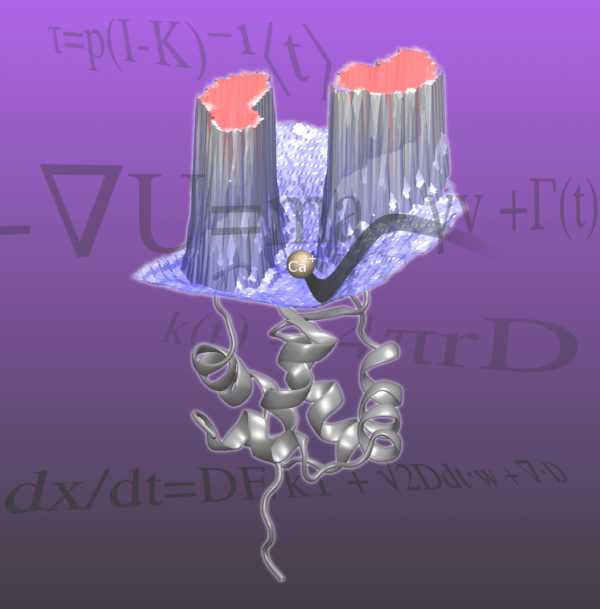
The kinetic rate constants of binding were estimated for four biochemically relevant molecular systems by a method that combines Brownian dynamics simulations with more detailed molecular dynamics simulations using milestoning theory. The rate constants found using this method were in good agreement with experimentally and theoretically obtained values. We predicted the association rate of a small charged molecule toward both a charged and an uncharged sphere and verified the estimated value with Smoluchowski theory. We also calculated the kon rate constant for superoxide dismutase with its natural substrate, O2-, in a validation of a previous experiment using similar methods but with a number of important improvements. We also calculated the kon for a new system: the N-terminal domain of Troponin C with its natural substrate Ca2+. The kon calculated for both systems closely resemble experimentally obtained values. This novel multiscale approach is computationally cheaper and more parallelizable compared to other methods of similar accuracy. We anticipate that this methodology will be useful for predicting kinetic rate constants and for understanding the process of binding between a small molecule and a protein receptor.
This data contains the input files, output files, trajectories, and gathered statistics from this investigation with the intent to allow other researchers to use this data to test their own models. - Date Collected
- 2013 to 2015
- Date Issued
- 2015
- Authors
- Note
-
Funding information: This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1144086. This work was funded in part by the National Institutes of Health (NIH) through the NIH Director’s New Innovator Award Program DP2-OD007237 and through the NSF XSEDE Supercomputer resources grant RAC CHE060073N to REA. Additional funding from the National Biomedical Computation Resource, NIH P41 GM103426, is gratefully acknowledged. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
- Topics
Formats
View formats within this collection
- Related Resources
- Votapka LW, Amaro RE (2015) Multiscale Estimation of Binding Kinetics Using Brownian Dynamics, Molecular Dynamics and Milestoning. PLoS Computational Biology 11(10): e1004381. https://doi.org/10.1371/journal.pcbi.1004381
Primary associated publication
Software
 Library Digital Collections
Library Digital Collections